Muscles in Action
Columbia University
ICCV 2023
Muscles in Action
Human motion is created by, and constrained by, our muscles. We take a first step at building computer vision methods that represent the internal muscle activity that causes motion. We present a new dataset, Muscles in Action (MIA), to learn to incorporate muscle activity into human motion representations. The dataset consists of 12.5 hours of synchronized video and surface electromyography (sEMG) data of 10 subjects performing various exercises. Using this dataset, we learn a bidirectional representation that predicts muscle activation from video, and conversely, reconstructs motion from muscle activation. We evaluate our model on in-distribution subjects and exercises, as well as on out-of-distribution subjects and exercises. We demonstrate how advances in modeling both modalities jointly can serve as conditioning for muscularly consistent motion generation. Putting muscles into computer vision systems will enable richer models of virtual humans, with applications in sports, fitness, and AR/VR.
Predicting Muscle Activation from Motion
We first model the forward direction: predicting muscle activation from motion (3D skeletons). Below are two qualitative results. The dashed line is the ground-truth and the solid line is our prediction. We adjusted the scale of the axis to fit the range of the ground truth muscle activation.
Predicting Motion from Muscle Activation
Next, we predict motion from muscle activation. Below are four qualitative results, where green represents our prediction and red is ground truth.
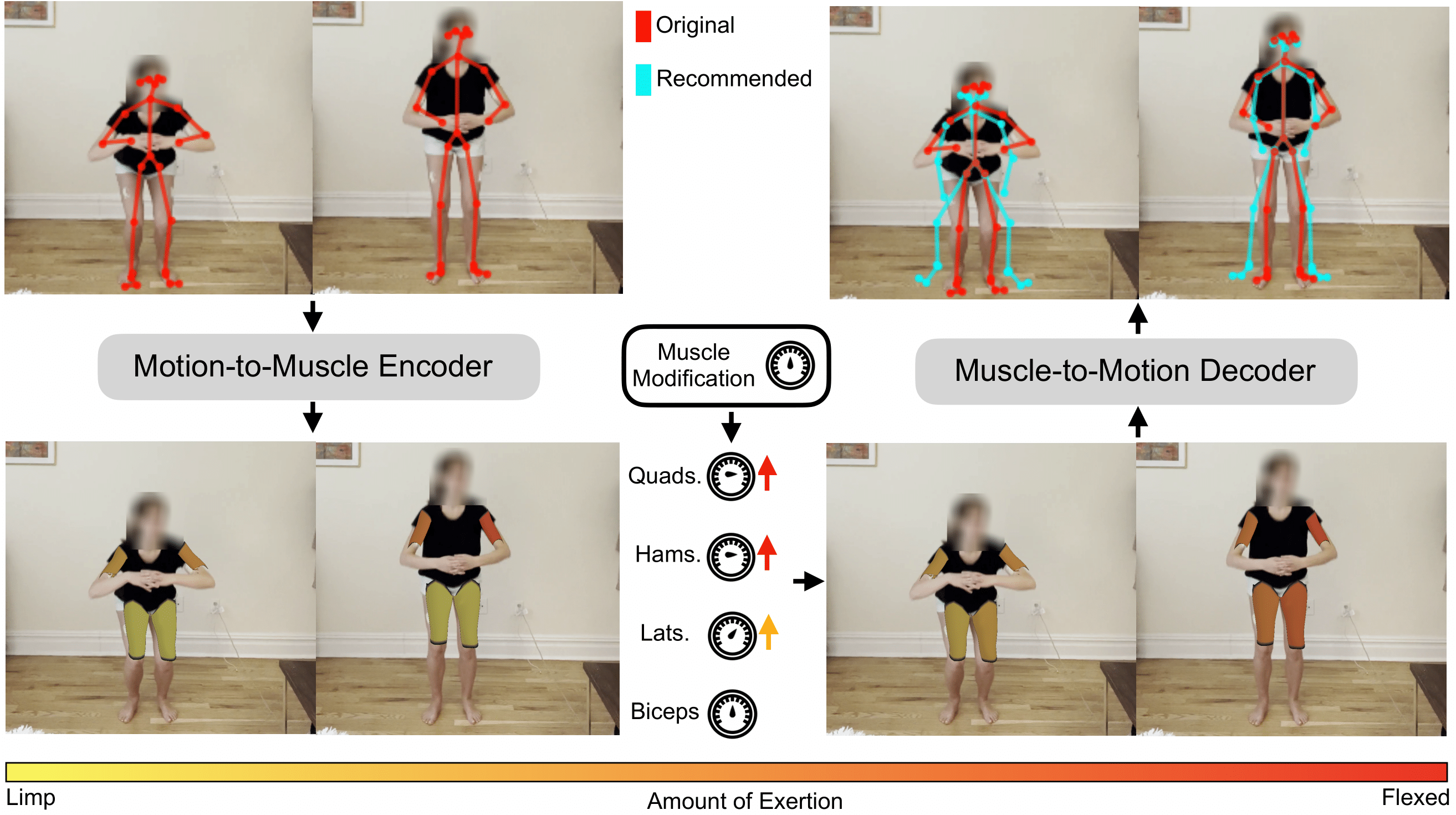
Muscle-Conditioned Motion Editing: Muscle Scaling
The first kind of muscle-conditioned motion editing is what we call muscle scaling. Given a motion, we predict the muscle activation from the input motion. We then scale a particular muscle group, which we then decode back into motion. This enables applications in which we want to modify a given exercise to target or avoid particular muscle groups.
The first video has a squat as the motion input. We scale the predicted hamstring+lateral activations up with a scaler greater than 1 before decoding to motion.
The second video has a rond-de-jambe as the motion input. We scale the predicted lateral activations up with a scaler greater than 1 before decoding to motion.
The third video has a skater as the motion input. We scale the predicted hamstring activations down with a scaler less than 1 before decoding to motion.
The fourth video has a side lunge as the motion input. We scale the predicted lateral activations up with a scaler greater than 1 before decoding to motion.
The second video has a rond-de-jambe as the motion input. We scale the predicted lateral activations up with a scaler greater than 1 before decoding to motion.
The third video has a skater as the motion input. We scale the predicted hamstring activations down with a scaler less than 1 before decoding to motion.
The fourth video has a side lunge as the motion input. We scale the predicted lateral activations up with a scaler greater than 1 before decoding to motion.
Muscle-Conditioned Motion Editing: Muscle Stitching
The second kind of muscle-conditioned motion editing that we show is what we call "muscle stitching", a reference to the classic computer vision problem of "image stitching". Given an input motion, we predict the muscle activation, and we replace the prediction for a particular muscle group with the predicted muscle activation for an entirely different motion. The sequence of muscle activations is now a blend from two different motions, which we then decode back into motion. This hybrid muscle activation has never been seen before in the training set, yet the decoded motion corresponds to the hybrid motions. These results illustrate our model's compositionality capabilites.
The first video has a front punch as the motion input, but we replace the predicted hamstring muscle activation with the hamstring prediction for a back kick.
The second video has a back kick as the motion input, but we replace the predicted bicep muscle activation with the bicep prediction for a front punch.
The third video has a front punch as the motion input, but we replace the predicted muscle activation for the quads with the quad prediction for a side lunge.
The fourth video has a side lunge as the motion input, but we replace the predicted muscle actiation for the biceps with the bicep prediction for a front punch.
The second video has a back kick as the motion input, but we replace the predicted bicep muscle activation with the bicep prediction for a front punch.
The third video has a front punch as the motion input, but we replace the predicted muscle activation for the quads with the quad prediction for a side lunge.
The fourth video has a side lunge as the motion input, but we replace the predicted muscle actiation for the biceps with the bicep prediction for a front punch.
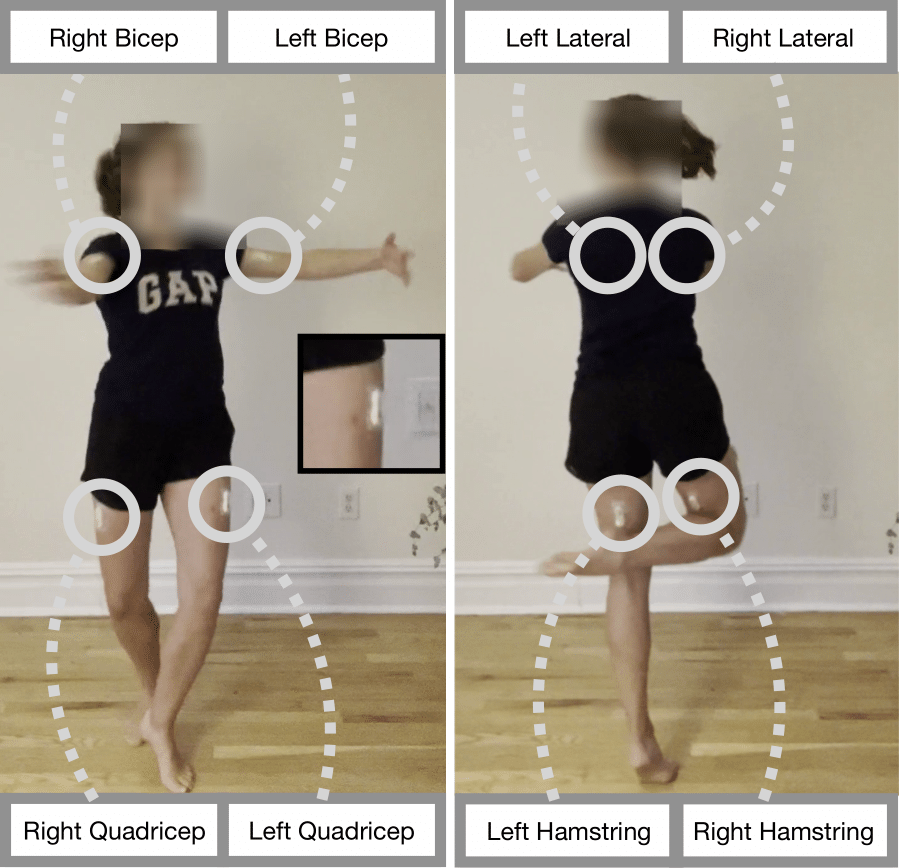
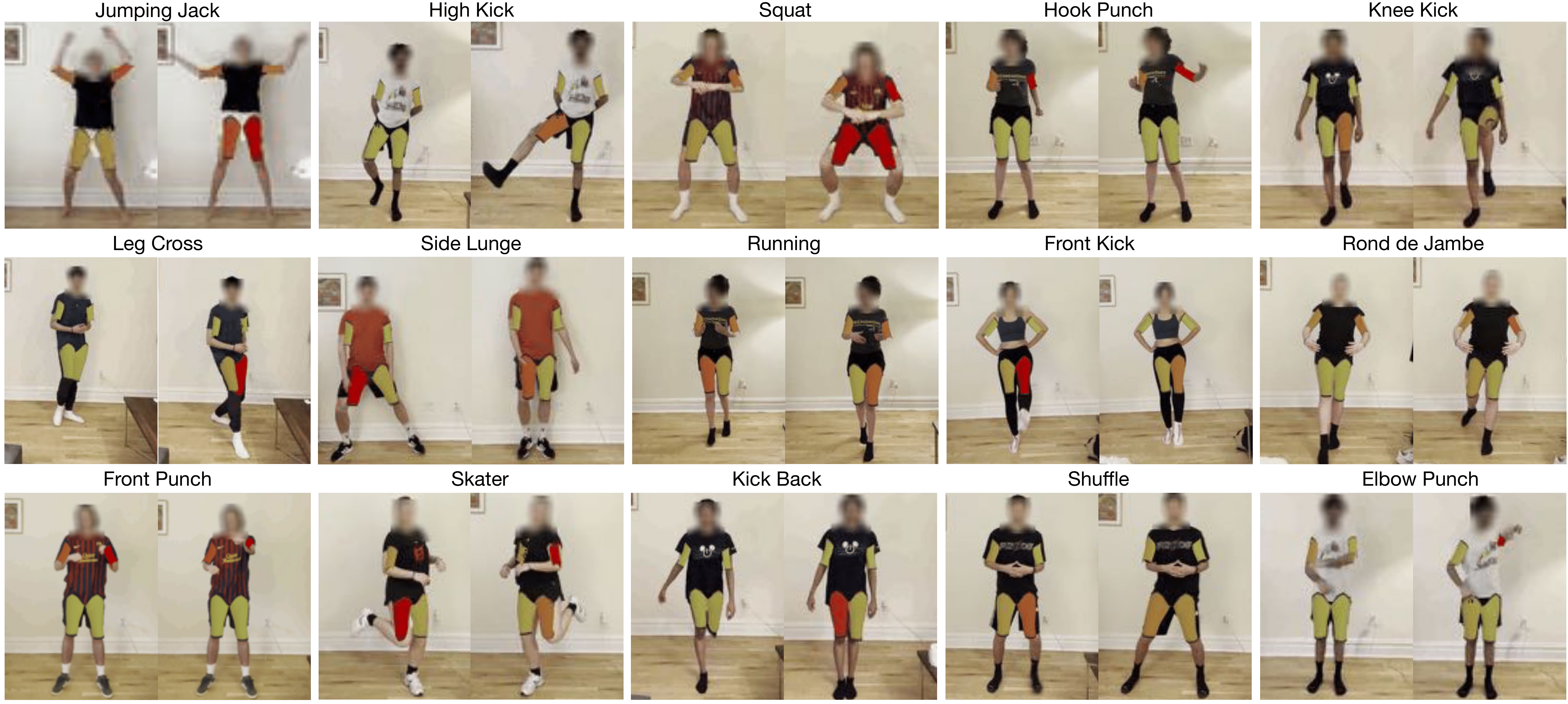
The Muscles in Action (MIA) Dataset
The MIA (Muscles in Action) dataset consists of 12.5 hours of synchronized video and sEMG signals, for eight muscles, across 10 subjects. These eight muscles
include the left and right biceps brachii (biceps), the left and
right latissimus dorsi (laterals), both quadriceps (quads),
and both biceps femoris (hamstrings). The collected sEMG
values correspond to the neuromuscular junction’s total bioelectric energy. The dataset consists of 15 exercises shown below. Each subject performed each exercise for 5 minutes,
and we asked them to vary the execution’s speed, effort,
and orientation. Across the 10 subjects in the
dataset, 5 are females and 5 are males.
We collected 75 minutes of data for each subject.
Code, Pre-trained Models, MIA Dataset (to be released soon):
Paper:
Acknowledgements
We would like to thank our subjects for participating in the dataset. We’d also like to thank Jianbo Shi, Georgia Gkioxari, Huy Ha and Kamyar Ghasemipour for their helpful feedback. This research is based on work partially supported by the NSF NRI Award #1925157. M.C. is supported by the Amazon CAIT PhD fellowship. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the sponsors